Facilitating Trustworthy Data Science
 When a data analysis result is published or shared, how do we know whether it is trustworthy? Conceptually, we must show that each step of the analysis (and each set of parameters used in the step) was justified, and that the original data was itself trustworthy.
When a data analysis result is published or shared, how do we know whether it is trustworthy? Conceptually, we must show that each step of the analysis (and each set of parameters used in the step) was justified, and that the original data was itself trustworthy.
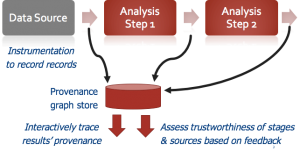
A team of Penn CIS and biomedical researchers are developing solutions for arbitrary data science computations: we instrumenting the analysis code to capture all of the computational stages, input records, and output values used during analysis — this is termed data provenance. We then learn how much to trust each computational stage and input file, in part by getting feedback from human experts, and in part by looking at whether the same stages and input files were used to produce scientifically valid answers for other computations. When mistakes are made, we provide debugging techniques and tools that use provenance to “trace back” faulty answers to the responsible inputs and code. The work is being deployed in high-throughput gene sequencing and neuroscience data analysis tasks here at Penn. More on the project is available at PennProvenance.NET, and the tools are being deployed in IEEG.org, a Penn-developed neuroscience data portal highlighted by NIH Director Francis Collins in a blog post.
For more information, see PennProvenance.NET and the Penn Database Group.
Faculty
- Zachary G. Ives
- Susan B. Davidson
- Val Tannen
- Sampath Kannan
- Junhyong Kim (Biology, Secondary in CIS)
- Brian Litt (Bioengineering and Neurology)
Sponsorship
Funded by grants from the NIH and NSF.
Students
- Nan Zheng
- Soonbo Han
- Yi Zhang
- Leshang Chen
- Rishabh Gupta (MSE)
- Prateek Singhal (MSE)